Mechanisms of Drug Sensitivity and Resistance in Small Cell Lung Cancer
Principal investigator: Roman Thomas, Christian Reinhardt
Project manager/Main research: Graciela Bosco
Researchers: Prof. Dr. med. Reinhard Büttner
, Univ.-Prof. Dr. med. Matthias Fischer, Dr. rer. nat. Julie George, PD Dr. rer. nat. Cécile Gouttefangeas, Univ.-Prof. Dr. rer. nat. Holger Grüll
, Dr. med. Grit S. Herter-Sprie, Univ.-Prof. Dr. med. Michael Hölzel, Univ.-Prof. Dr. rer. nat. Stefan Knapp
,Univ.-Prof. Dr.-Ing. Ulrich Lang, Univ.-Prof. Dr. rer. nat. Manolis Pasparakis, Univ.-Prof. Dr. rer. nat. Martin Peifer, Dr. rer. nat. M. Nieves Peltzer
, Univ.-Prof. Dr. med. Alexander Quaas, Univ.-Prof. Dr. rer. nat. Hans-Georg Rammensee
, Univ.-Prof. Dr. med. H. Christian Reinhardt, Prof. Dr. med. Ugur Sahin, Dr. med.Anne M. Schultheis, Univ.-Prof. Dr. med. Dr. rer. nat. Michal R. Schweiger, Univ.-Prof. Dr. med. Martin L. Sos, Univ.-Prof. Dr. med. Roman K. Thomas
, Dr. rer. nat. Silvia von Karstedt, Prof. Dr. rer. nat. Henning Walczak, Univ.-Prof. Dr. med. Jürgen Wolf
, PD Dr. rer. nat. F. Thomas Wunderlich
Title of research project: „CRC1399 - Mechanisms of Drug Sensitivity and Resistance in Small Cell Lung Cancer”
Funding: DFG
Project partners: University of Cologne, University Hospital Cologne, Max Planck Institute for Metabolism Research, TRON Translational Oncology at the University Medical Center of the Johannes Gutenberg University Mainz, University of Bonn, Goethe University Frankfurt, University of Tübingen, CIO Centrum für Integrierte Onkologie
Project endurance: (07.2019 – 07.2023)
Project area: Medizin (205)
Cluster: CHEOPS
Software: Own developments, irods, blat, samtools, bwamem, sclust, u.a.
Introduction
Small cell lung cancer (SCLC), with up to 8000 new diagnosed cases in Germany each year, is the most aggressive lung cancer subtype showing rapid cell division, treatment resistance and tendency to metastasize early. Recently, the first significant improvement in decades has been seen with the addition of immunotherapy to chemotherapy, but the effects are very modest. Much more exploration is needed to identify predictive biomarkers (i.e., biomolecules that can be used for early detection, diagnostic and prognostic) and to understand the underlying molecular mechanisms that cause this devastating disease. Building on recent discoveries in SCLC genetics and transcriptomics, we launch an orchestrated pursuit of mechanistic analyses, with the specific emphasis on identifying druggable features of SCLC like those underlying drug sensitivity, drug resistance and poor patience survival. We aim at advancing the understanding of the molecular development of the disease with the ultimate goal of transferring our novel discoveries into clinical applications that improve the survival of SCLS patients.
Methods
We are a highly interdisciplinary consortium covering expertise in biochemistry and signaling, structural biology, medical chemistry and structure-guided drug design, cancer immunology and modeling, computational cancer genomics, molecular pathology, as well as clinical trials. Our methods range from very well established techniques to novel transdisciplinary approaches, among them: generation of new SCLC models, comparative sequencing studies of drug-sensitive and drug-resistant SCLC tumors and functional characterization of these mutations, complex computational algorithms applied on whole-exome sequencing of SCLS specimens to establish the temporal order of mutational events, mass cytometry and a novel multiplexed immunohistochemistry platform to probe the tumor microenvironment before and after the occurrence of drug resistance, all of these together with long-term monitoring of patients in the clinic.
Results
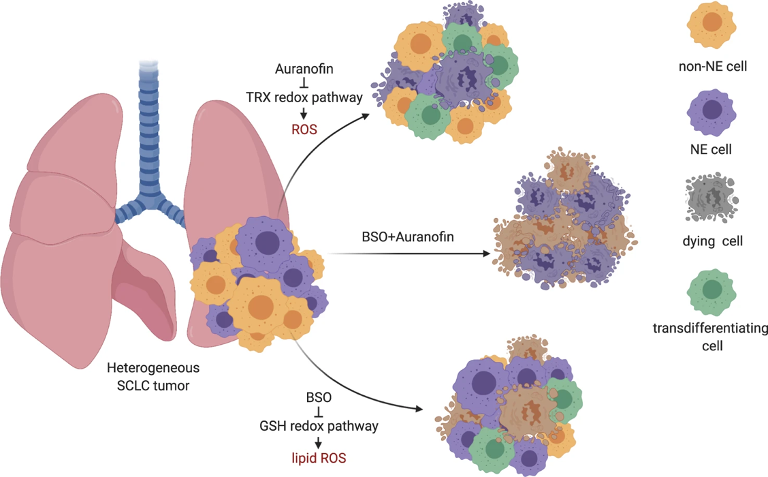
There are different molecular subtypes of SCLC that can differ in their responses to various treatments. Heterogeneous tumors, that contain two or more of these subtypes, show the presence of neuroendocrine (NE) cells (NE), and non-neuroendocrine (non-NE) cells together. Recently, members of our Team, in an extended collaboration, discovered that the natural death of SCLC tumor cells can be activated in non-NE-cells using BSO (which induces ferroptosis: iron-dependent cell death caused by oxidative stress), while in the other type, the death of NE cells can be induced by Auranofin (an inhibitor of the TRX anti-oxidant pathway). Interestingly, when applying the treatment individually using only one of these mechanisms, the threatened tumor cells transformed themselves (so called, transdifferentiating cells) to avoid being killed (see Fig.1). Therefore, dual targeting, i.e., tackling both pathways simultaneously, has higher therapeutic efficiency in this kind of tumors.
Discussion
Unfortunately, many other mechanisms that could potentially be used to induce natural tumor-cell death are already suppressed when the cancer is detected in patients. Therefore, characteristic tumor signatures that are potentially targetable or can serve for prediction or prognostic are very valuable. In general, throughout its life, any cell is exposed to various sources of DNA damage (like UV radiation or tobacco) that result in mutations. These mutational processes leave characteristic fingerprints in the DNA of cancer cells that can be exploited. Members of our Team have developed a new tool, called CaMuS, that allow us to disentangle and discover cancer mutational signatures in the DNA of SCLC cancer cells. The algorithm initially fits known “reference signatures” to the DNA data and then performs non-matrix factorization on the unexplained data to infer potentially novel patterns of mutational processes. After being derived mathematically, these mutation signatures have to be experimentally validated. Such studies in metastatic cancer, like SCLC, can provide important insights into therapeutic resistance mechanisms that can lead the development of novel treatments.
Outlook
Combining different approaches from modelling to flow cytometry and proteomics, from sequencing to mathematical algorithms, from laboratory probes to clinical trials, will continue generating new insights into SCLC biology and providing entry routes for targeted therapeutic intervention that derive in the improvement of life-quality and survival of SCLC patients.
Ferroptosis response segregates small cell lung cancer (SCLC) neuroendocrine subtypes
Christina M. Bebber, Emily S. Thomas, Jenny Stroh, Zhiyi Chen, Ariadne Androulidaki, Anna Schmitt, Michaela N. Höhne, Lukas Stüker, Cleidson de Pádua Alves, Armin Khonsari, Marcel A. Dammert, Fatma Parmaksiz, Hannah L. Tumbrink, Filippo Beleggia, Martin L. Sos, Jan Riemer, Julie George, Susanne Brodesser, Roman K. Thomas, H. Christian Reinhardt & Silvia von Karstedt
Nature Communications. 2021 Apr 6, volume 12,
Article number: 2048 (2021)
Characterizing genetic intra-tumor heterogeneity across 2,658 human cancer genomes
Stefan C.Dentro, Ignaty Leshchiner, Kerstin Haase, Maxime Tarabichi, Jeff Wintersinger, Amit G.Deshwar, KaixianYu, Yulia Rubanova, Geoff Macintyre, Jonas Demeulemeester, Ignacio Vázquez-García, Kortine Kleinheinz, Dimitri G.Livitz, Salem Malikic, Nilgun Donmez, Subhajit Sengupta, Pavana Anur, Clemency Jolly, Marek Cmero, Daniel Rosebrock, Steven E.Schumacher, Yu Fan, Matthew Fittall, Ruben M.Drews, Xiaotong Yao, Thomas B.K.Watkins, Juhee Lee, Matthias Schlesner, Hongtu Zhu, David J.Adams, Nicholas McGranahan, Charles Swanton, Gad Getz, Paul C.Boutros, MarcinImielinski, Rameen Beroukhim, S. Cenk Sahinalp, Yuan Ji, Martin Peifer, Inigo Martincorena, Florian Markowetz, Ville Mustonen, Ke Yuan, Moritz Gerstung, Paul T.Spellman, Wenyi Wang, Quaid D.Morris, David C.Wedge, Peter Van Loo
Cell. 2021 Apr 7. ScienceDirect.
In Press
Integrin Activation Enables Sensitive Detection of Functional CD4 + and CD8 + T Cells: Application to Characterize SARS-CoV-2 Immunity
Anna Schöllhorn, Juliane Schuhmacher, Luciana Besedovsky, Rolf Fendel, Anja T R Jensen, Stefan Stevanović, Tanja Lange, Hans-Georg Rammensee, Jan Born, Cécile Gouttefangeas, Stoyan Dimitrov
Front Immunol, 2021 Mar 29;12:626308. doi: 10.3389/fimmu.2021.626308
PMID: 33854501
CD4+ T Cells: Multitasking Cells in the Duty of Cancer Immunotherapy
Jennifer R Richardson, Anna Schöllhorn, Cécile Gouttefangeas, Juliane Schuhmacher
Cancers (Basel). 2021 Feb 3;13(4):596.doi: 10.3390/cancers13040596
PMID: 33546283
Deep Learning Predicts HPV Association in Oropharyngeal Squamous Cell Carcinomas and Identifies Patients with a Favorable Prognosis Using Regular H&E Stains
Klein S, Quaas A, Quantius J, Löser H, Meinel J, Peifer M, Wagner S, Gattenlöhner S, Wittekindt C, von Knebel Doeberitz M, Prigge ES, Langer C, Noh KW, Maltseva M, Reinhardt HC, Büttner R, Klussmann JP, Wuerdemann N.
Clin Cancer Res. 2021 Feb 15;27(4):1131-1138. doi: 10.1158/1078-0432.CCR-20-3596. Epub 2020 Dec 1
PMID: 33262137
Active Akt signaling triggers CLL toward Richter transformation via overactivation of Notch1
Kohlhaas V, Blakemore SJ, Al-Maarri M, Nickel N, Pal M, Roth A, Hövelmeyer N, Schäfer SC, Knittel G, Lohneis P, Nikolic M, Wiederstein JL, Franitza M, Georgomonolis T, Reinart N, Herling M, Herling C, Hartmann EM, Rosenwald A, Klapper W, Büttner R, Moia R, Rossi D, Boldorini R, Gaidano G, Frenzel LP, Reinhardt HC, Brüning JC, Hallek M, Krüger M, Peifer M, Pallasch CP, Wunderlich FT.
Blood. 2021 Feb 4;137(5):646-660. doi: 10.1182/blood.2020005734.
PMID: 33538798
Texture analysis of iodine maps and conventional images for k-nearest neighbor classification of benign and metastatic lung nodules
Lennartz S, Mager A, Große Hokamp N, Schäfer S, Zopfs D, Maintz D, Reinhardt HC, Thomas RK, Caldeira L, Persigehl T.
Cancer Imaging. 2021 Jan 26;21(1):17. doi: 10.1186/s40644-020-00374-3.
PMID: 33499939
An Autochthonous Mouse Model of Myd88- and BCL2-Driven Diffuse Large B-cell Lymphoma Reveals Actionable Molecular Vulnerabilities
Flümann R, Rehkämper T, Nieper P, Pfeiffer P, Holzem A, Klein S, Bhatia S, Kochanek M, Kisis I, Pelzer BW, Ahlert H, Hauer J, da Palma Guerreiro A, Ryan JA, Reimann M, Riabinska A, Wiederstein J, Krüger M, Deckert M, Altmüller J, Klatt AR, Frenzel LP, Pasqualucci L, Béguelin W, Melnick AM, Sander S, Montesinos-Rongen M, Brunn A, Lohneis P, Büttner R, Kashkar H, Borkhardt A, Letai A, Persigehl T, Peifer M, Schmitt CA, Reinhardt HC, Knittel G.
Blood Cancer Discov. 2021 Jan;2(1):70-91. doi: 10.1158/2643-3230.BCD-19-0059.
PMID: 33447829
Adaptive T-cell immunity controls senescence-prone MyD88- or CARD11-mutant B-cell lymphomas.
Reimann M, Schrezenmeier JF, Richter-Pechanska P, Dolnik A, Hick TP, Schleich K, Cai X, Fan DNY, Lohneis P, Masswig S, Denker S, Busse A, Knittel G, Flümann R, Childs D, Childs L, Gätjens-Sanchez AM, Bullinger L, Rosenwald A, Reinhardt HC, Schmitt CA.
Blood. 2020 Nov 24:blood.2020005244. doi: 10.1182/blood.2020005244.
PMID: 33232972
CaMuS: simultaneous fitting and de novo imputation of cancer mutational.
Cartolano M, Abedpour N, Achter V, Yang TP, Ackermann S, Fischer M, Peifer M. signature.
Sci Rep. 2020 Nov 9;10(1):19316. doi: 10.1038/s41598-020-75753-8.
PMID: 33168834
Acute myeloid leukemia-induced remodeling of the human bone marrow niche predicts clinical outcome.
Chen Y, Hoffmeister LM, Zaun Y, Arnold L, Schmid KW, Giebel B, Klein-Hitpass L, Hanenberg H, Squire A, Reinhardt HC, Dührsen U, Bertram S, Hanoun M.
Blood Adv. 2020 Oct 27;4(20):5257-5268. doi: 10.1182/bloodadvances.2020001808.
PMID: 33108453
Economic Impact of the Introduction of Outpatient Medical Specialist Care (ASV) of Gastrointestinal Cancer Patients from a German Hospital Management Perspective.
Jakobs F, Drost RMWA, Kron A, Heinen J, Hallek M, Reinhardt HC, Zander T, Kron F.
Oncol Res Treat. 2020;43(10):498-505. doi: 10.1159/000509618. Epub 2020 Sep 21
PMID: 32957103
Landscape of G-quadruplex DNA structural regions in breast cancer.
Hänsel-Hertsch R, Simeone A, Shea A, Hui WWI, Zyner KG, Marsico G, Rueda OM, Bruna A, Martin A, Zhang X, Adhikari S, Tannahill D, Caldas C, Balasubramanian S.
Nat Genet. 2020 Sep;52(9):878-883. doi: 10.1038/s41588-020-0672-8. Epub 2020 Aug 3.
PMID: 32747825
Integrin activation enables simultaneous and sensitive detection of functional virus-specific CD4+ and CD8+ T cells.
Anna Schöllhorn, Juliane Schuhmacher, Luciana Besedovsky, Rolf Fendel, Anja T.R. Jensen,
Stefan Stevanović, Tanja Lange, Hans-Georg Rammensee, Jan Born, Cécile Gouttefangeas, Stoyan Dimitrov
Research Square
DOI: 10.21203/rs.3.rs-67111/v1
Integrative Analysis of Pleomorphic Dermal Sarcomas Reveals Fibroblastic Differentiation and Susceptibility to Immunotherapy.
Klein S, Quaas A, Noh KW, Cartolano M, Abedpour N, Mauch C, Quantius J, Reinhardt HC, Buettner R, Peifer M, Helbig D.
Clin Cancer Res. 2020 Aug 17. doi: 10.1158/1078-0432.CCR-20-1899. Epub ahead of print.
PMID: 32817080
Precision medicine in non-small cell lung cancer: Current applications and future directions.
Yang SR, Schultheis AM, Yu H, Mandelker D, Ladanyi M, Büttner R.
Semin Cancer Biol. 2020 Jul 27:S1044-579X(20)30164-4. doi: 10.1016
PMID: 32730814 Review
Dissecting intratumour heterogeneity of nodal B-cell lymphomas at the transcriptional, genetic and drug-response levels.
Roider T, Seufert J, Uvarovskii A, Frauhammer F, Bordas M, Abedpour N, Stolarczyk M, Mallm JP, Herbst SA, Bruch PM, Balke-Want H, Hundemer M, Rippe K, Goeppert B, Seiffert M, Brors B, Mechtersheimer G, Zenz T, Peifer M, Chapuy B, Schlesner M, Müller-Tidow C, Fröhling S, Huber W, Anders S, Dietrich S.
Nat Cell Biol. 2020 Jul;22(7):896-906. doi: 10.1038/s41556-020-0532-x. Epub 2020 Jun 15.
PMID: 32541878
New Approaches to SCLC Therapy: From the Laboratory to the Clinic.
John T Poirier, Julie George, Taofeek K Owonikoko, Anton Berns, Elisabeth Brambilla, Lauren A Byers, David Carbone, Huanhuan J Chen, Camilla L Christensen, Caroline Dive, Anna F Farago, Ramaswamy Govindan, Christine Hann, Matthew D Hellmann, Leora Horn, Jane E Johnson, Young S Ju, Sumin Kang, Mark Krasnow, James Lee, Se-Hoon Lee, Jonathan Lehman, Benjamin Lok, Christine Lovly, David MacPherson, David McFadden, John Minna, Matthew Oser, Keunchil Park, Kwon-Sik Park, Yves Pommier, Vito Quaranta , Neal Ready, Julien Sage, Giorgio Scagliotti, Martin L Sos, Kate D Sutherland, William D Travis, Christopher R Vakoc, Sarah J Wait, Ignacio Wistuba, Kwok Kin Wong, Hua Zhang, Jillian Daigneault, Jacinta Wiens, Charles M Rudin, Trudy G Oliver.
J Thorac Oncol. 2020 Apr;15(4):520-540. doi: 10.1016/j.jtho.2020.01.016
PMID: 32018053
Fighting Against Promoter DNA Hyper-Methylation: Protective Histone Modification Profiles of Stress-Resistant Intestinal Stem Cells
Torsten Thalheim, Lydia Hopp, Maria Herberg, Susann Siebert, Christiane Kerner, Marianne Quaas, Michal R Schweiger, Gabriela Aust, Joerg Galle.
Int J Mol Sci. 2020 Mar 12;21(6):1941.
PMID: 32178409
ATM activity in T cells is critical for immune surveillance of lymphoma in vivo.
Riabinska A, Lehrmann D, Jachimowicz RD, Knittel G, Fritz C, Schmitt A, Geyer A, Heneweer C, Wittersheim M, Frenzel LP, Torgovnick A, Wiederstein JL, Wunderlich CM, Ortmann M, Paillard A, Wößmann W, Borkhardt A, Burdach S, Hansmann ML, Rosenwald A, Perner S, Mall G, Klapper W, Merseburg A, Krüger M, Grüll H, Persigehl T, Wunderlich FT, Peifer M, Utermöhlen O, Büttner R, Beleggia F, Reinhardt HC.
Leukemia. 2020 Mar;34(3):771-786. doi: 10.1038/s41375-019-0618-2. Epub 2019 Nov 5.
PMID: 31690822
The evolutionary history of 2,658 cancers.
Gerstung M, Jolly C, Leshchiner I, Dentro SC, Gonzalez S, Rosebrock D, Mitchell TJ, Rubanova Y, Anur P, Yu K, Tarabichi M, Deshwar A, Wintersinger J, Kleinheinz K, Vázquez-García I, Haase K, Jerman L, Sengupta S, Macintyre G, Malikic S, Donmez N, Livitz DG, Cmero M, Demeulemeester J, Schumacher S, Fan Y, Yao X, Lee J, Schlesner M, Boutros PC, Bowtell DD, Zhu H, Getz G, Imielinski M, Beroukhim R, Sahinalp SC, Ji Y, Peifer M, Markowetz F, Mustonen V, Yuan K, Wang W, Morris QD; PCAWG Evolution & Heterogeneity Working Group, Spellman PT, Wedge DC, Van Loo P; PCAWG Consortium.
Nature. 2020 Feb;578(7793):122-128. doi: 10.1038/s41586-019-1907-7. Epub 2020 Feb 6.
PMID: 32025013
MYC paralog-dependent apoptotic priming orchestrates a spectrum of vulnerabilities in small cell lung cancer.
Dammert MA, Brägelmann J, Olsen RR, Böhm S, Monhasery N, Whitney CP, Chalishazar MD, Tumbrink HL, Guthrie MR, Klein S, Ireland AS, Ryan J, Schmitt A, Marx A, Ozretić L, Castiglione R, Lorenz C, Jachimowicz RD, Wolf E, Thomas RK, Poirier JT, Büttner R, Sen T, Byers LA, Reinhardt HC, Letai A, Oliver TG, Sos ML.
Nat Commun. 2019 Aug 2;10(1):3485. doi: 10.1038/s41467-019-11371-x.
PMID: 31375684
Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data.
Rudin CM, Poirier JT, Byers LA, Dive C, Dowlati A, George J, Heymach JV, Johnson JE, Lehman JM, MacPherson D, Massion PP, Minna JD, Oliver TG, Quaranta V, Sage J, Thomas RK, Vakoc CR, Gazdar AF.
Nat Rev Cancer. 2019 May;19(5):289-297. doi: 10.1038/s41568-019-0133-9